Katelyn Arnold is a Research Assistant Professor in the Division of Chemical Biology and Medicinal Chemistry (CBMC). Dr. Arnold’s research interest is in therapeutic development of synthetic heparin. She uses chemoenzymatically synthetized oligosaccharides in various animal models to investigate the relationship between oligosaccharide structure and function to understand anti-inflammatory mechanisms. This work also involves pharmacokinetic studies using methods and standards specific for synthetic heparin.
Drug Discovery: PhD Program
Uniquely Positioned to Bridge the Chemical and Biological Worlds
 Chemical Biology and Medicinal Chemistry (CBMC) is a dynamic, multifaceted graduate program dedicated to improving human health through research leading to new concepts for the design and development of therapeutic and diagnostic agents.
Chemical Biology and Medicinal Chemistry (CBMC) is a dynamic, multifaceted graduate program dedicated to improving human health through research leading to new concepts for the design and development of therapeutic and diagnostic agents.
Our program seamlessly blends chemistry and biology, which distinguishes it from traditional graduate programs. We develop and exploit novel chemical tools relevant to the fields of biochemistry, biology, pharmacology, and medicine. Research is directed toward biomedical and pharmaceutical discovery by applying both chemical and biological principles to interactions between molecular structure and biological activity.
All of our graduates have been successful in finding desirable positions. An important measure of the success of our program is whether students are obtaining their first-choice postdoctoral positions. To a large extent, our recent graduates are receiving multiple offers from top labs. Our recent graduates have taken postdoctoral positions in prestigious labs at Harvard, Duke, Scripps, and MIT, to name a few.
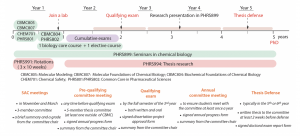
Students enter the CBMC program through application to either the Pharmaceutical Sciences program or the Biological and Biomedical Sciences Program, a large umbrella program offering an entry to 16 PhD programs on the UNC-Chapel Hill campus. The Figure below shows the overview of the CBMC program. To complete the CBMC graduate program, a student needs to pass 8 courses, cumulative exams, qualifying exam and thesis defense. In addition, students will participate in the weekly seminar series throughout the training. Over the last 10 years, more than 50 PhD students have graduated from the CBMC program and the average time to completion is approximately 5 years. More details are described in the Student Handbook.
CBMC 807: Molecular Foundations in Chemical Biology
Prerequisites: Students are expected to have a solid understanding of introductory organic chemistry as taught at the undergraduate level.
This course provides a review of important concepts in organic chemistry as they apply to biological research. Topics include a review of intermolecular interactions as they apply to biological structures and function, a discussion of how small molecules interact with their targets, an overview of synthetic methods that relate particularly to drug molecules, and basic strategies of drug design.
CBMC 805: Molecular Modeling
Prerequisites: None
This course provides a general introduction to the field of Molecular Modeling while providing relevant applications of theory to both academic and industrial research endeavors.
PHRS 801: Common Core in Pharmaceutical Sciences
Prerequisites: None.
This course provides an interdisciplinary environment for students from each of the four Divisional PhD programs in UNC Eshelman School of Pharmacy. Students will learn about and develop skills in topics related to responsible conduct of research, pharmaceutical development, professional development, and independent development.
PHRS 899: Seminars in Chemical Biology
Students must register for PHRS 899 each semester of their graduate program. However, only 4 credits of PHRS 899 (2 credits for MS) may count toward requirement for the PhD degree. Seminars are conducted jointly with the Division of Chemical Biology and Bioorganic Chemistry in the Department of Chemistry. Attendance at all Division seminars is mandatory and two unexcused absences will result in a grade of F. As an additional component of the seminar requirements of the graduate curriculum, attendance by all students is required during a student’s Doctoral Defense. These Defense seminars are held outside of the normal seminar series.
Each student is required to present a seminar in PHRS 899 either in the student’s third year or in the beginning of his/her fourth year and will be considered in assigning the grade in PHRS 899. In this seminar, the student critically reviews the area pertinent to his/her thesis topic making sure that s/he discusses studies that include his/her group’s contributions and those of other laboratories and includes a description of the student’s ongoing studies that add to this body of research. Faculty members will evaluate the student seminar. Students receiving an overall failing evaluation on the presentation will receive an “incomplete” grade in PHRS 899 for that semester and must consult with the seminar coordinator before giving a make-up seminar at a later date. Additionally, students receiving “incomplete” grades may be advised to seek further training in presentations. In those cases where the student’s research contains intellectual property (IP) and where disclosure risks the IP, a student can provide a comprehensive review of a different subject in medicinal chemistry. Permission to do so will require approval of the thesis adviser and the seminar coordinator. Each seminar topic, title, summary, and research article must be approved by the student’s research adviser and seminar coordinator. A student presenter should send the title of their seminar talk to the Graduate Program Coordinator upon request.
PHRS 991: Research in Pharmaceutical Sciences (Research Rotations)
During the first two semesters, the student conducts three ~10-week research rotation projects, each under the supervision of a different faculty member. These rotation projects are considered as course work for PHRS 991. Students select rotations from any of the CBMC Faculty. With approval of the DDGS, students may also perform rotation projects with faculty outside the labs of the CBMC Faculty, especially when the rotation will provide training in an area that is outside of the expertise of the CBMC Faculty.
To select an adviser for each research rotation, the student should interview members of the CBMC faculty about possible projects. Prior to each rotation, the student will turn in a RESEARCH ROTATION LAB SELECTION FORM (Appendix A) to the Graduate Program Coordinator. Over the course of the first year each student is encouraged to schedule individual interviews with all members of the CBMC faculty. Although varied slightly from year to year, the rotations usually start in the late August and end in the late April in the next year. The schedule for the 2022-2023 academic year is as follows:
Fall: Monday, August 22 – Friday, October 28 (OK to start before August 22)
Winter: Monday, November 7-Friday, January 20
Spring: Monday, January 30 – Friday, April 7
Students admitted in the fall semesters can begin their rotations the summer before. In that scenario, the student must contact the Graduate Program Coordinator at least four weeks prior to the start of the rotation. The summer rotation will be considered as one of their three required rotations. A waiver for one research rotation may be granted if a student has previously completed substantial independent research. Students seeking such a waiver must petition the DDGS, and provide information (e.g. reports, manuscripts, grant proposals, and/or letters from research advisers) about their previous research. If a student has obtained a specific fellowship to work with a CBMC faculty member, then research rotations may be optional.
During lab rotations, students are expected to work in the laboratory at least 20 hours per week. Students are fully integrated into the laboratory during their rotation projects and are involved in lab meetings and journal clubs. At the beginning and end of each rotation, the rotation adviser and student review a written or oral statement of expectations for the student’s performance in the laboratory. These discussions provide the student with the advisers’ expectations and critical comments on areas of excellence and weakness. Guidelines for the research rotations are described in the PHRS 991 syllabus.
At the end of each rotation, students will submit a written report using the Research in Pharmaceutical Sciences – Student Evaluation Form (Appendix B) to the Graduate Program Coordinator. Students will also present the results of their rotation projects to their Student Advisory Committee (SAC). The SAC committee, in consultation with the rotation adviser, will provide a brief summary and evaluation and submit a grade to the Graduate Program Coordinator and the DDGS to be entered at the end of the semester. After the third rotation, students will present the work of their rotation in the CBMC End of Year Mini-Symposium.
The DDGS serves as the temporary advisor for the first-year students who enroll in PHRS 991. The SAC committee provides additional mentoring and consists of three CBMC faculty. The SAC is formed at the beginning of the first semester based on a student’s request and availability of faculty members.
CHEM 701 (Introduction to Laboratory Safety)
Prerequisites: first year graduate student status or permission of instructor
This course provides an overview of safety rules and regulations, guidance in safe laboratory practice, and creates a culture of laboratory safety.
CBMC 804A: Biochemical Foundations of Chemical Biology.
Prerequisites: CHEM 466, BIOC 505, 601, or PHCO 643; or permission of instructors.
This course covers core biochemical and molecular biology techniques, concepts, and tools used to conduct research at the interface of chemistry and biology. Topics include enzymology, characterization of drug-target interactions, mechanisms-based inhibitor design, assay design and development, targeting kinases and GPCRs, biopharmaceuticals, gene therapy, nucleic-acid binding agents, information-based drugs, chemical tools to study epigenetics, harnessing biosynthetic pathways for chemical diversity, and other recent advances and techniques in drug discovery.
CBMC 804B: Foundations of Chemical Biology Journal Club.
Prerequisites: Enrollment in CBMC 804A.
This course is a series of presentations by students that run in concert with CBMC 804A.
Biology Core Course
Each student has the option to choose one 3- or 4-credit hour course on campus that is focused on biological systems or techniques. A good starting point to find such a course is the BBSP website. Some examples include PHCO701 (Introduction to Molecular Pharmacology), BIOC706 (Biochemistry of Human Disease), GNET631 (Advanced Molecular Biology), CBIO643 (Cell Structure and Function) and CBIO893 (Advanced Cell Biology).
PHRS 802: Drug Development and Professional Skills Development
Prerequisites: None.
This course provides an interdisciplinary environment for students from each of the four Divisional PhD programs in UNC Eshelman School of Pharmacy. Students will learn about the general process of drug development and develop associated professional skills.
Elective Course: Students have the option to take one elective course of their interest. There is no requirement on the number of credit hours of the course. Students typically choose a course that provides specific skills and knowledge their thesis work needs.
PHRS 994: Research in Pharmaceutical Sciences (Thesis Research)
The students begin to register 3 credit hours PHRS 994 each semester once they have chosen the thesis adviser. Guidelines for the thesis research are described in the PHRS 994 syllabus.
CBMC Faculty
Jeff Aubé
ACCEPTING DOCTORAL STUDENTS
The Aubé laboratory uses synthetic chemistry to enable the study of biological pathways and as starting points for drug discovery. Current efforts in the group include the study of new opioids lacking side effects, new approaches for the treatment of tuberculosis, androgen biosynthesis inhibitor discovery, the search for RNA-protein interaction inhibitors, and the development of new synthetic methods.
Alison Axtman
The Axtman lab is devoted to characterization of the dark proteome, especially those proteins with underexplored roles in the brain. Our interests lie at the interface of chemistry and biology, with a focus on using small molecules, specifically potent and selective chemical probes, to explore and impact disease-propagating pathways. Active projects are aimed at finding pre-clinical small molecule candidates that, after further optimization, can help address the need for new therapeutics in human diseases. Our scientists are working to design and synthesize chemical modulators and develop screening assays. All data and reagents are openly shared to facilitate and expedite scientific advancement.
Albert Bowers
ACCEPTING DOCTORAL STUDENTS
Albert Bowers received his PhD in organic chemistry (synthetic methods) from the University of Illinois at Chicago. He carried out postdoctoral research (total synthesis) at Colorado State University before moving as an NIH sponsored fellow to Harvard Medical School (biosynthesis). He is a member of the UNC Lineberger Comprehensive Cancer Center and affiliate member of the Center for Integrative Chemical Biology and Drug Discovery.
Rafael Couñago
Rafael M. Couñago, PhD, is a Research Associate Professor in the Division of Chemical Biology and Medicinal Chemistry Department in the UNC Eshelman School of Pharmacy and a Principal Investigator at the Structural Genomics Consortium (SGC) at UNC. Rafael´s research group at SGC-UNC uses protein biochemistry, structural biology and cell-based assays to illuminate protein function and explore new therapeutic strategies for human diseases.
David Drewry
ACCEPTING DOCTORAL STUDENTS
The Drewry lab in focused on designing, synthesizing, evaluating, and sharing small molecule chemical probes for protein kinases. These tools are used to build a deeper understanding of disease pathways and facilitate identification of important targets for drug discovery. Through wide ranging partnerships with academic and industrial groups, the Drewry lab is building a Kinase Chemogenomic Set (KCGS) that is available to the community for screening.
Kevin Frankowski
Research in the Frankowski lab uses synthetic chemistry to develop new approaches for the treatment of unmet medical needs. Our current efforts focus on programs to treat metastatic cancer, hepatitis C virus infection and the development of chemical tools for studying dopamine and sigma receptors.
Stephen Frye
Dr. Stephen Frye is currently a Professor in the Center for Integrative Chemical Biology and Drug Discovery (CICBDD) which he previously directed at the University of North Carolina at Chapel Hill (UNC). Prior to joining UNC to create the CICBDD in 2007, Dr. Frye was the world-wide vice president of Discovery Medicinal Chemistry (DMC) at GlaxoSmithKline (GSK). Dr. Frye led DMC for seven years, overseeing five departments and more than 200 chemists in the U.S. and U.K. developing global protein target-class chemical science for GSK. During his 20-year career at GSK, the teams led by Dr. Frye successfully developed three FDA approved drugs: Avodart, a dual 5a-reductase inhibitor for treatment of benign prostatic hyperplasia, Tykerb, a dual erbB2/EGFR inhibitor for the treatment of metastatic breast cancer, and Pazopanib, a multi-targeted receptor tyrosine kinase inhibitor for the treatment of renal cell carcinoma and soft tissue sarcoma. As founding director and current faculty member in the CICBDD at UNC, Dr. Frye plays a key role in translational research through collaborative drug discovery projects with other UNC faculty. A clinical candidate from one of these projects created in the Center is now progressing through multiple human trials. In addition, his lab has established a leading program in the area of chemical biology of chromatin regulation with an emphasis on protein-protein interactions dependent upon lysine methylation. Dr. Frye has published more than 130 papers in the fields of organic and medicinal chemistry.
Lauren Haar
Our projects focus on investigating the role that refined spatial and temporal control of intracellular signaling cascades can play in the progression of cardiovascular injury. We use a research strategy involving plasmid and optogenetic protein engineering, high content screening, high resolution microscopy and physiologically based cell analysis. With this approach we hope to uncover new targets for therapeutic development by better defining signaling cascades that drive cardiovascular disease response.
Nate Hathaway
ACCEPTING DOCTORAL STUDENTS
The Hathaway lab was established at UNC with a founding idea that the group could make a contribution to understanding dynamic epigenetic processes by using unique chemical biology approaches they pioneered. Through the combination of protein bioengineering, synthetic organic chemistry, and mammalian cell-based model systems, they have created platforms that use chemically tethered enzymatic recruitment to specific chromatin loci to produce a host of mechanistic insights. The Hathaway group also has drug discovery programs to identify new small molecules that inhibit disease relevant epigenetic pathways both for research purposes and as potential future therapeutics.
Lindsey Ingerman James
ACCEPTING DOCTORAL STUDENTS
The James lab is interested in modulating the activity of chromatin reader proteins with small-molecule ligands, specifically potent and selective chemical probes, in order to open new avenues of research in the field of chromatin biology and potentially translate to compounds of therapeutic value. They are also interested in applying novel probe-based techniques, such as affinity labeling technologies, to the study of epigenetic regulators.
Michael Bruce Jarstfer
Michael Jarstfer, PhD, is an Associate Professor within the Division of Chemical Biology and Medicinal Chemistry and the Associate Dean for Graduate Education. He is also the Director of Graduate Studies for the Pharmaceutical Sciences PhD program. Dr. Jarstfer has expertise in drug target identification, high-throughput-screening, medicinal chemistry, and compound optimization for drug discovery as well as pharmacology in preclinical animal models.
David S. Lawrence
NOT ACCEPTING GRADUATE STUDENTS
The Lawrence lab works to understand the biochemical processes of the cell by studying them as they happen in the cell as opposed to studying them in vitro. He currently focuses on applying his discoveries to cancer detection and treatment and, to a more limited extent, inflammatory diseases.
Andrew L Lee
ACCEPTING DOCTORAL STUDENTS
Andrew Lee studies the role of conformational dynamics in protein function, conformational changes, enzyme catalysis, drug binding, and allostery. His laboratory uses a variety of biophysical and biochemical tools, especially NMR spectroscopy. NMR spectroscopy is a powerful approach that yields atomic-resolution molecular information and is uniquely sensitive to molecular fluctuations over a broad range of timescales.
Jian Liu
ACCEPTING DOCTORAL STUDENTS
Research in the Jian Liu group is focused on glycobiology and glycobiochemistry, an emerging field that emphasizes the biological functions of carbohydrates. We are particularly interested in understanding the biosynthetic mechanism of sulfated polysaccharides known as heparan sulfate and heparin.
Rihe Liu, PhD
ACCEPTING DOCTORAL STUDENTS
The Liu laboratory’s research interests focus on the development and application of novel drug target-binding affinity molecules by integrating directed molecular selection and evolution, ligand design and engineering, in vitro cellular and signaling characterization, and in vivo therapeutic efficacy studies in tumor mouse models. The Liu laboratory has extensive experiences in the design, synthesis, characterization, and delivery of diagnostic and therapeutic agents based on both polypeptides and polynucleotides.
Robert McGinty
ACCEPTING DOCTORAL STUDENTS
The McGinty lab studies molecular mechanisms of chromatin signaling. By pairing atomic precision protein chemistry with high resolution structural biology, they aim to understand how the nucleosome functions as a signaling hub for gene expression, DNA replication, and DNA damage repair in development and disease.
Eugene Muratov
Dr. Muratov served as a corresponding author on an approach used by regulators to initially screen new chemical products for toxic effects. They have proposed an improvement that could increase the accuracy of toxicity estimation to as much as 85 percent, saving millions of dollars and years of development time for new drugs and other products while improving safety.
Samantha Pattenden
ACCEPTING DOCTORAL STUDENTS
The Pattenden lab develops innovative techniques in chromatin-based therapeutic target discovery and cancer diagnostics. Our research program enables discovery of novel molecular targets, pathways and mechanisms. Our central strategy exploits tumor-specific changes in chromatin accessibility, a universal feature that is directly linked with transcriptional activation, DNA damage repair, replication, RNA processing, and nuclear organization.
Kenneth Pearce, Jr
Ken Pearce, Ph D is the director of the Center for Integrative Chemical Biology and Drug Discovery. Pearce’s primary expertise and interests are fundamentals of protein methods, biochemical and cell assay development, medium- and high-throughput screening, hit validation and mode-of-action, biophysical methods for characterizing protein-protein and small molecule-protein interactions, and structure-activity relationships for early drug discovery. He joined the center as director of lead discovery and characterization in mid-2015 after spending over 18 years at GlaxoSmithKline and legacy companies in the Molecular Discovery Research organization.
Konstantin Popov
The Popov Lab develops inventive, cutting-edge approaches to solve problems in modern computational structural biology and drug discovery. Their computational research, in collaboration with experimental screening and medicinal chemistry efforts in the Center for Integrative Chemical Biology and Drug Discovery enables the identification of novel chemical probes and drug candidates to advance understanding of biological processes. Some of their recent projects include:
• Identification and characterization of allosteric and cryptic binding sites
• Development of AI-driven methods for accelerated virtual screening (VS)
• AI approaches for DNA-encoded library (DEL)-guided virtual screening (VS) and new-generation DEL library designs
• Collaborative hit discovery and optimization projects
Paul Sapienza
Paul Sapienza is a research assistant professor in the Division of Chemical Biology and Medicinal Chemistry at the UNC Eshelman School of Pharmacy. His research aims to further understanding of the role of dynamics in biomolecular recognition, enzymatic catalysis, and allostery. He uses nuclear magnetic resonance spectroscopy to study protein dynamics on multiple timescales, while other tools such as calorimetry, crystallography, and kinetics serve to link dynamics with function. He is focusing on thymidylate synthase as it is an enzyme with a multistep catalytic cycle, is a cancer drug target, and exhibits negative cooperativity (allostery).
Scott Singleton
Scott Singleton is engaged in educational innovation and research. His work attempts to build on what is understood about memory and attention to devise, test, and implement effective teaching and learning strategies. His current work focuses on teaching that positively affects student engagement in the classroom, identifying core basic science concepts that serve as threshold concepts for pharmacy students, and evaluating the transfer of learning between courses in the professional PharmD program.
Junjiang Sun
Junjiang Sun is a Research Assistant Professor in the Division of Chemical Biology and Medicinal Chemistry (CBMC). Sun’s research interests focus on gene therapy for hemophilia, hemophilia associated joint disease (Hemophilia Arthropathy), by expressing bypassing agents (activated coagulation factor V, IX) via AAV vectors. This technology platform provides novel therapeutic approaches for rheumatoid arthritis (RA) and osteoarthritis (OA).
Alexander Tropsha, PhD
ACCEPTING DOCTORAL STUDENTS
Alex Tropsha, Ph.D., is an expert in the fields of computational chemistry, cheminformatics and data science. His laboratory develops new methodologies, software tools and applications in the areas of computer-assisted drug design, chemical toxicology, materials informatics, text mining, and health care informatics.
Xiaodong Wang
The Wang lab is interested in developing drug leads/candidates for kinase, phosphate kinase and protein targets identified by UNC faculty and external investigators. We have successfully used the structure- and/or ligand-based drug design approaches to deliver compounds to clinic (MerTK inhibitors such as MRX-2843) or licensing (IDH1 inhibitor, co-developed with NCATs). We will continue to apply the similar approaches for drug discovery towards new targets.
Tim Willson
The Willson laboratory is home to the US site of the SGC, an open science consortium that accelerates research on the lesser studied regions of the genome. The laboratory works closely with pharma companies and academic investigators to develop small molecule chemical probes for hundreds of dark kinases that are openly shared with the scientific community. Current research has led to the development of the Kinase Chemogenomic Set (KCGS) that contains selective inhibitors of more than 200 kinases as well as high quality chemical probes for several of the dark kinases.
Yongmei Xu
Dr. Xu co-authored the Heparin study with Dr. Jian Liu and Dr. Lindhardt. Heparin is a naturally occurring polysaccharide that prevents blood clotting, or coagulation, and has been in use since the late 1930s. A polysaccharide is a long chain of carbohydrate molecules.
Qisheng Zhang
ACCEPTING DOCTORAL STUDENTS
The Zhang lab studies lipid signaling pathways that are involved in human disease by developing novel chemical probes and technologies. They currently focus on discovering new bioactive lipids, developing small molecule modulators and biosensors for lipid metabolizing enzymes, and applying their research results to novel diagnosis and treatment of cancer, Parkinson’s disease, and antimicrobial resistance.
CBMC Joint Appointments
Jeff Aubé
ACCEPTING DOCTORAL STUDENTS
The Aubé laboratory uses synthetic chemistry to enable the study of biological pathways and as starting points for drug discovery. Current efforts in the group include the study of new opioids lacking side effects, new approaches for the treatment of tuberculosis, androgen biosynthesis inhibitor discovery, the search for RNA-protein interaction inhibitors, and the development of new synthetic methods.
David S. Lawrence
NOT ACCEPTING GRADUATE STUDENTS
The Lawrence lab works to understand the biochemical processes of the cell by studying them as they happen in the cell as opposed to studying them in vitro. He currently focuses on applying his discoveries to cancer detection and treatment and, to a more limited extent, inflammatory diseases.
Robert McGinty
ACCEPTING DOCTORAL STUDENTS
The McGinty lab studies molecular mechanisms of chromatin signaling. By pairing atomic precision protein chemistry with high resolution structural biology, they aim to understand how the nucleosome functions as a signaling hub for gene expression, DNA replication, and DNA damage repair in development and disease.
CBMC Emeriti Faculty
CBMC Adjunct Faculty
Yuriy Abramov, Ph.D
Nikolay Dokholyan, PhD, MS
Sean Ekins, Ph.D.
Denis Fourches, PhD
Clark D Jeffries, Ph.D.
Jian Jin, Ph.D.
Kyoko Nakagawa-Goto, Ph.D.
David Nichols, Ph.D.
Lars Pedersen, Ph.D.
pederse2@niehs.nih.gov
Adjunct Associate Professor, UNC Eshelman School of Pharmacy
Staff Scientist, Head of Collaborative Crystallography Lab, NIEHS
Jun Tang
Dr. Tang is a collaborating member from ViiV Healthcare with the UNC-Chapel Hill HIV Cure Center. His research is focused on discovering novel HIV latency reversal agents with better tolerability and exploring new ways to clear the latent T cells upon activation and viral particles.
Lan Xie, Ph.D.
Weifan Zheng, Ph.D.
CBMC Staff
 The Division of Chemical Biology and Medicinal Chemistry is training the next generation of researchers specializing in drug discovery, and a clear strength is the cadre of young scientists comprising our graduate students and postdoctoral fellows. As of the fall 2016 semester, there were 24 graduate students and 26 postdoctoral fellows working as students, teachers and scholars within the division.
The Division of Chemical Biology and Medicinal Chemistry is training the next generation of researchers specializing in drug discovery, and a clear strength is the cadre of young scientists comprising our graduate students and postdoctoral fellows. As of the fall 2016 semester, there were 24 graduate students and 26 postdoctoral fellows working as students, teachers and scholars within the division.
Our students receive full financial support, including a competitive stipend, paid tuition and health insurance. They complete the Ph.D. program in approximately five years, and our graduates have taken postdocs in prestigious labs (at Harvard, Duke, Scripps and MIT to name a few) and found desirable positions in academia and pharmaceutical and biotechnology companies. We meet many of our students for the first time during the School’s Recruitment Weekend each January.
Download Brochure
-
 Ikeer Mancera Ortiz
Ikeer Mancera Ortiz
-
 Brian Anderson
Brian Anderson
-
 Aasif Ansari
Aasif Ansari
-
 Michael Arcidiacono
Michael Arcidiacono
-
 Jon-Michael Beasley
Jon-Michael Beasley
-
 Matthew Bowler
Matthew Bowler
-
 Peter Buttery
Peter Buttery
-
 Jacob Capener
Jacob Capener
-
 Jenna Colton
Jenna Colton
-
 Glory Dan-Dukor
Glory Dan-Dukor
-
 Raymond Flax
Raymond Flax
-
 Emily Flory
Emily Flory
-
 Merrill Froney
Merrill Froney
-
 Avery Huber
Avery Huber
-
 Rebecca Johnson
Rebecca Johnson
-
 Jiwoong Lim
Jiwoong Lim
-
 Lilyan Mather
Lilyan Mather
-
 Amanda McGowan
Amanda McGowan
-
 Eric Merten
Eric Merten
-
 Han Wee Ong
Han Wee Ong
-
 Jarrett Pelton
Jarrett Pelton
-
 Jixuan Qiao
Jixuan Qiao
-
 Meghan Ricciardi
Meghan Ricciardi
-
 Madison Routh
Madison Routh
-
 Devan Shell
Devan Shell
-
 David Shirley
David Shirley
-
 Benjamin Strickland
Benjamin Strickland
-
 Holli-Joi Sullivan
Holli-Joi Sullivan
-
 Michelle Thomas
Michelle Thomas
-
 Ashley Trojniak
Ashley Trojniak
-
 Jessica Danielle Umana
Jessica Danielle Umana
-
 Sara Wasserman
Sara Wasserman
-
 James Wellnitz
James Wellnitz
-
 Xindi Zhang
Xindi Zhang
-
 Yani Zhao
Yani Zhao
-
 Ivanna Zhilinskaya
Ivanna Zhilinskaya







































