Nate Hathaway Lab
Nate Hathaway Lab
 In multicellular organisms, genes must be regulated with high precision during stem-cell differentiation to achieve normal development. Pathologically, the loss of proper gene regulation caused by defects in chromatin regulatory enzymes has been found to be a driving force in cancer initiation and progression.
In multicellular organisms, genes must be regulated with high precision during stem-cell differentiation to achieve normal development. Pathologically, the loss of proper gene regulation caused by defects in chromatin regulatory enzymes has been found to be a driving force in cancer initiation and progression.
The Hathaway lab uses a combination of chemical-biology and cell-biology approaches to unravel the molecular mechanisms that govern gene expression. We utilize new tools wielding an unprecedented level of temporal control to visualize changes in chromatin structure and function in mammalian cells and animal models. In addition, we seek to identify small molecule inhibitors that are selective for chromatin regulatory enzymes with the potential for future human therapeutics.
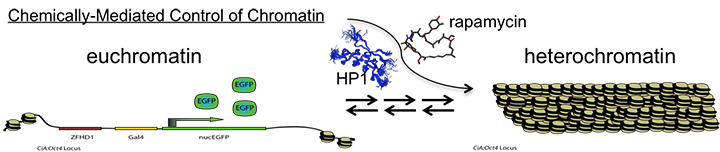
The Hathaway lab is positioned along the interface between chemical biology and cell biology. Our lab uses novel chemical tools and in vivo assay systems to gain insights on how genes are epigenetically regulated in mammalian cells. Specifically, we want to understand the key molecular mechanisms responsible for complex transformations of chromatin states such as transitions between active euchromatin and repressive heterochromatin. Most of our current work focuses on the HP1-mediated heterochromatin pathway and additionally how histone acetylation state influences gene transcription. We are also developing new chemically based gene control systems capable of allele specific regulation genome wide.
One arm of the Hathaway lab at UNC focuses on further understanding the transitions between active (euchromatin) and inactive (heterochromatin) genes, with a desire to quantify the dynamics of the enzymatic machinery acting on chromatin to cause heritable changes in gene expression. These epigenetic modifications have been increasingly implicated as having a key role in cancer initiation and progression. The overarching goal of this arm of the lab is to understand the basic mechanisms underlying these complex regulatory networks, which can lead to human disease when disrupted. We have conducted high-throughput screens to identify chemical inhibitors of heterochromatin formation and memory that could have application in the treatment of cancer. Additionally, we are generating and utilizing novel chromatin in vivo assays to ask questions about gene regulatory pathways not previously addressable. The new insights for heterochromatin propagation and fidelity generated by this work will lead to generalizable models with applications to a broad range of chromatin regulatory pathways.
The second arm of the Hathaway lab uses advances we have made in chemical chromatin control systems to apply new epigenome editing technology genome wide with allele specific precision. We accomplish this by using deactivated CRISPR associated protein 9 (dCas9) to engage any desired mammalian genomic loci by specific promoter- and gene-targeting guide RNAs. We couple this technology with chemical recruitment strategies, providing the ability to recruit epigenetic regulators to sites bound by dCas9 in a dose dependent and reversible manner. This technology unearthed new insights into how chromatin landscape influences chromatin regulatory pathways. We are now pioneering this technology to control disease relevant genes in human cancer as a new therapeutic approach in collaboration with clinician scientists.
Developing projects in the lab include:
- Identification of the molecular determinants of heterochromatin formation
- Mechanisms leading to heterochromatin memory through cellular generations
- High-throughput screens to identify small molecule inhibitors of epigenetic regulators and use of these inhibitors to gain new mechanistic insights
- Controlling cellular state with novel synthetic epigenetic regulators
- New chemically-based approaches to understanding and treating human cancer
Research Keywords:
- Epigenetics
- Chromatin Biology
- Chemical Biology
- Cell Biology
- Cancer Research
- Gene Regulation
- Bioengineering
- Drug Discovery
10 Most Recent Lab Publications:
Chiarella AM, Butler KV, Gryder BE, Lu D, Wang TA, Yu X, Pomella S, Khan J, Jin J#, Hathaway NA#. Dose-dependent activation of gene expression is achieved using CRISPR and small molecules that recruit endogenous chromatin machinery. Nat Biotechnol. 2019 Nov 11.
Madigan VJ, Yuziuk JA, Chiarella AM, Tyson TO, Meganck RM, Elmore ZC, Tse LV, Hathaway NA, Asokan A. Ring finger protein 121 is a potent regulator of adeno-associated viral genome transcription. PLoS Pathog. 2019 Aug 6;15(8):e1007988.
Vignaux PA, Bregio C, Hathaway NA#. Contribution of promoter DNA sequence to heterochromatin formation velocity and memory of gene repression in mouse embryo fibroblasts. PLoS One. 2019;14(7):e0217699.
MacDonald IA, Butler KV, Herring LE, Clinkscales SE, Yelagandula R, Stecher K, Bell O, Graves LM, Jin J, Hathaway NA#. Pathway-Based High-Throughput Chemical Screen Identifies Compounds That Decouple Heterochromatin Transformations. SLAS Discov. 2019 May 30;:2472555219849838.
Headley KM, Kedziora KM, Alejo A, Lai EZ, Purvis JE, Hathaway NA#. Chemical screen for epigenetic barriers to single allele activation of Oct4. Stem Cell Res. 2019 May 24;38:101470
Khodaverdian V, Tapadar S*, MacDonald IA*, Xu Y, Ho PY, Bridges A, Rajpurohit P, Sanghani BA, Fan Y, Thangaraju M, Hathaway NA#, Oyelere AK#. Deferiprone: Pan-selective Histone Lysine Demethylase Inhibition Activity and Structure Activity Relationship Study. Sci Rep. 2019 Mar 18;9(1):4802.
Chory EJ, Calarco JP, Hathaway NA, Bell O, Neel DS, Crabtree GR. Nucleosome Turnover Regulates Histone Methylation Patterns over the Genome. Mol Cell. 2019 Jan 3;73(1):61-72.e3.
Chiarella AM, Wang TA, Butler KV, Jin J, Hathaway NA#. Repressing Gene Transcription by Redirecting Cellular Machinery with Chemical Epigenetic Modifiers. J Vis Exp. 2018 Sep 20;(139).
Chiarella AM*, Quimby AL*, Mehrab-Mohseni M, Velasco B, Kasoji SK, Davis IJ, Dayton PA, Hathaway NA#, Pattenden SG#. Cavitation Enhancement Increases the Efficiency and Consistency of Chromatin Fragmentation from Fixed Cells for Downstream Quantitative Applications. Biochemistry. 2018 May 15;57(19):2756-2761.Butler KV*, Chiarella AM*, Jin J#, Hathaway NA#. Targeted Gene Repression Using Novel Bifunctional Molecules to Harness Endogenous Histone Deacetylation Activity. ACS Synth Biol. 2018 Jan 19;7(1):38-45.
Select publications prior to UNC
Hathaway NA*, Bell O*, Hodges C, Miller EL, Neel DS, Crabtree GR. Dynamics and Memory of Heterochromatin in Living Cells. Cell. 2012 Jun 22;149(7):1447-60. See Preview in Cell and Highlight in Nature Methods and Nature Reviews Genetics. Recommended by Faculty of 1000.
Kirkpatrick DS*, Hathaway NA*, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006 Jul;8(7):700-10. Recommended by Faculty of 1000.
Link to Full Publication list
http://www.ncbi.nlm.nih.gov/sites/myncbi/nathaniel.hathaway.1/bibliography/40595033/public/
Notes: * = equal contribution, # = Corresponding author
Collaborations
Modern science is driven by collaborative projects; we are always interested in pursuing new and interesting avenues in research. Potential collaborators should contact Nate Hathaway, PhD.
Graduate Students
The Hathaway Lab is part of two graduate training programs at UNC. Students first must gain admittance to either the CBMC or BBSP programs. Motivated perspective and first-year graduate students from either program are always encouraged to contact Nate Hathaway about potential rotation projects.
Postdoctoral Fellows
We are not actively seeking postdoctoral fellows at this time. However, talented scientists with a desire to train in the lab and a strong scientific track record should contact Nate Hathaway to inquire about potential opportunities
Jessica Danielle Umana
Xiaokang Yan
Patricia Vignaux (Graduate Student)
Anna Marie Chiarella (Graduate Student)
Ian MacDonald (Postdoctoral Fellow)
April Wang (Undergraduate Researcher)
Celyn Bregio (Undergraduate Researcher)
Aidin Alejo (Undergraduate Researcher)
Ellie Lai (Undergraduate Researcher)
Bryan Obika (Undergraduate Researcher)
Austin Sun (Undergraduate Researcher)
Spencer Maranto (Undergraduate Laboratory Assistant)
Everett Desern (Undergraduate Laboratory Assistant)
Sloan Walters (Undergraduate Laboratory Assistant)
Polina Cherkez (Undergraduate Laboratory Assistant)
Kristina Cala (High School Student)
Ben Wu (High School Student)
2018

2017

2016
The Hathaway Lab gratefully acknowledges funding from the following sources:
Laboratory Support Grants/Awards :
National Institute of General Medical Sciences (R01)
National Institute of General Medical Sciences (Supplemental Equipment Grant)
Eshelman Institute for Innovation (Tier 3 award, Tier 2 award)
Eshelman School of Pharmacy
AACP New Investigator Award
IBM Junior Faculty Development Award
University Cancer Research Fund, University of North Carolina at Chapel Hill
North Carolina Biotechnology Center Institutional Development Grant
Selected Student Awards :
NSF Graduate Research Fellowships Program (Katie Headley)
Eshelman Institute for Innovation Student Award (Anna Chiarella)
GMB NIH T-32 (Anna Chiarella)
MiBio nIH T-32 (Katie Headley)
NC TraCS Institute Grant (Anna Chiarella)
Keystone Meeting Travel Award (Anna Chiarella)




