Science
High-quality chemical probes are highly valuable reagents to explore biology of their target proteins. The most useful chemical probes meet five criteria:
-
The chemical probe is a potent inhibitor of its primary target, ideally with an in vitro potency in biochemical assays < 100 nM.
-
The chemical probe demonstrates selectivity over other proteins, with at least a 30-fold window over closely related targets in the same protein family.
-
The chemical probe shows activity in cellular assays at or below a concentration of 1 µM.
-
A close analog of the chemical probe that lacks activity on the primary target is available as a negative control compound.
-
The chemical probe is publicly available without restrictions on its use.
Dark Kinases
SGC-UNC has generated high quality chemical probes for several dark kinases.
SGC-AAK1-1 for adaptor protein 2-associated kinase (AAK1) and BMP-2 inducible kinase (BMP2K)
SGC-GAK-1 for cyclin G-associated Kinase (GAK)
SGC-CK2-1 for casein kinase 2 (CSNK2)
SGC-CLK-1 for Cdc2-like kinases 1, 2, and 4 (CLK1/CLK2/CLK4)
SGC-CAMKK2-1 for calcium/calmodulin-dependent protein kinase kinase 2 and 1 (CAMKK2/CAMKK1)
SGC-STK17B-1 for serine/threonine kinase 17B (STK17B)
We have also collaborated with SGC scientists in Frankfurt, Campinas, and Oxford to release chemical probes for several additional kinases.

IDG
The Illuminating the Druggable Genome (IDG) Program is an NIH-sponsored initiative to study the properties and functions of dark proteins within the known families of drug targets. During the IDG pilot phase the program developed a website, called Pharos, that integrates information about dark proteins so that researchers everywhere can easily access it, catalyzing their own research and helping them find new proteins that may be of interest. In addition, IDG supported several technology development projects to enable the study of dark proteins in a high throughput manner. During the Implementation Phase, IDG established a large network of collaborators to expand the informatics tools developed in the Pilot Phase, elucidate the function of understudied proteins from three key druggable protein families (GPCRs, ion channels, and kinases), and disseminate the IDG-generated resources to the greater scientific community.

Table: Classification of the 134 IDG dark kinases by substrate. Listed alphabetically within each category. Protein kinases: putative serine, threonine, and tyrosine kinases. Pseudokinases: same fold as the protein kinases, but lack one or more canonical catalytic site residues. Other kinases: includes inositol, glycerol, nucleotide, and thiamine kinases. Regulatory proteins: phosphotransferase activity not established; modulate other kinases.
SGC-UNC is contributing to the Drug and Resource Generation Center for Kinases led by Gary Johnson. We will identify cell active inhibitors for the dark kinases on the IDG list. Many of these kinase inhibitors will fill gaps in our public chemogenomic set KCGS. IDG data and resources are available at the Dark Kinase Knowledgebase.

IDG Workflow:
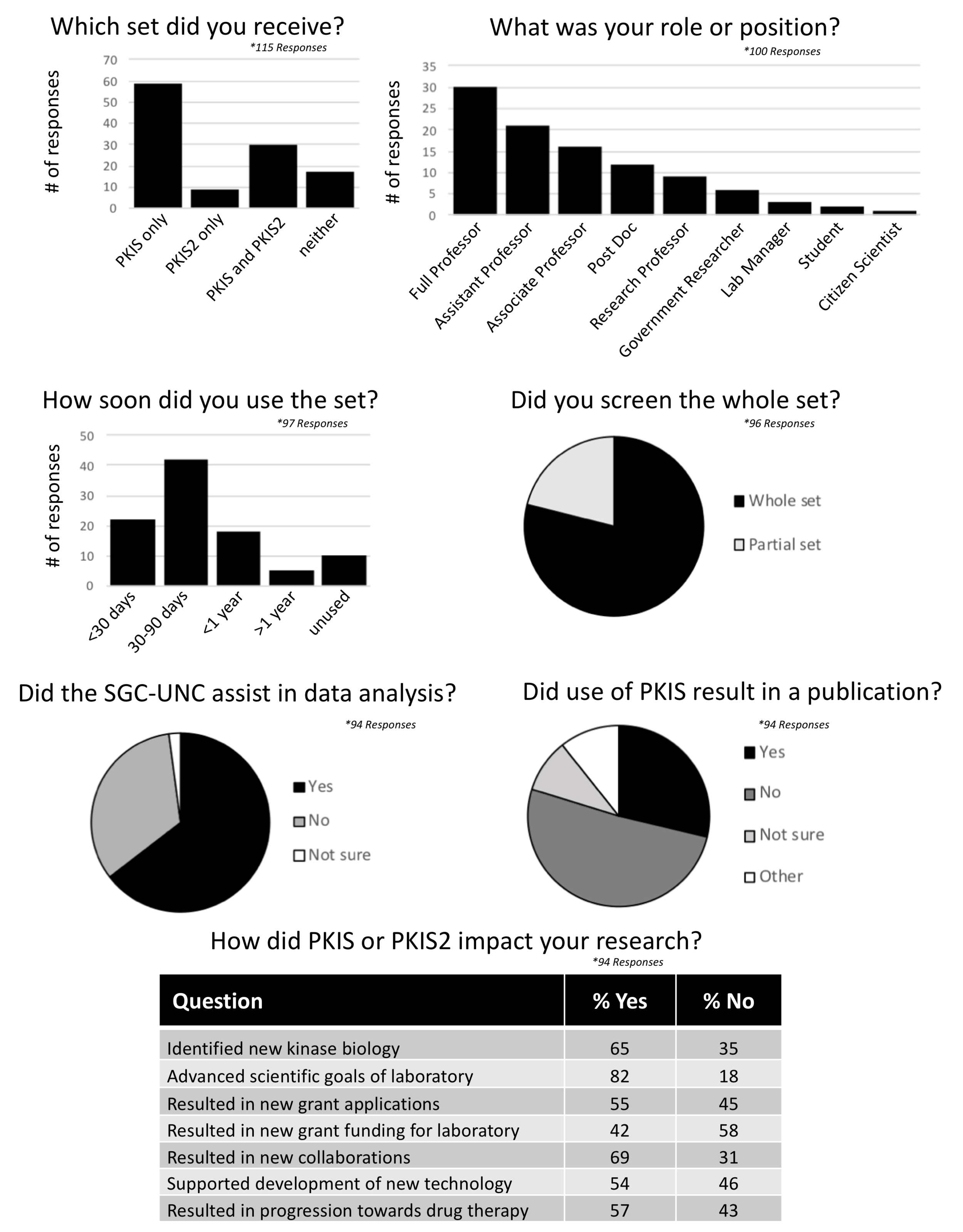
PKIS User Survey
PKIS and PKIS2 are kinase chemogenomic sets distributed by GSK between 2011-2015 and by the SGC-UNC from 2015-2017. The sets were shipped to over 300 laboratories. To gather data on the value of these publically-available chemogenomic sets as resources for open science, a survey was conducted on August 28th, 2018. The survey was designed by Maryann Feldman of the UNC Business School. 356 scientists who had received or requested a copy of PKIS or PKIS2 were contacted by email and 115 completed the survey.
A summary of their responses is listed below:
NanoBRET
Kinase inhibitor potency and selectivity is routinely measured using biochemical assays, which typically measure binding to the enzyme or inhibition of enzymatic activity in vitro. However, many compounds show a much lower potency for their inhibition of kinase activity in cells. This disconnect may arise from multiple factors, including the use of isolated kinase domains rather than full length protein, the absence of important partner proteins, or the use of ATP at concentrations well below what is found in living cells. For this reason, the SGC-UNC in collaboration with the Promega Corporation has applied NanoBRET technology to the development of kinase target engagement assays in live cells.
The development of nanoluciferase (Nluc) by Keith Wood and colleagues has accelerated interest in the use of bioluminescence to monitor target engagement of small molecules with kinases and other proteins. Nluc is an extremely bright luciferase derived from the deep-sea shrimp Oplophorus gacsilorstis that was generated by a combination of protein engineering and a chemical optimization of the enzyme substrate to be both significantly smaller and 150-fold brighter than firefly or Renilla luciferases. When employed with an efficient red-shifted fluorophore acceptor, the combination of greater light intensity and wide spectral resolution gives improved detection sensitivity and dynamic range over conventional BRET technologies. The high intensity requires the protein-Nluc fusion to be transiently expressed at only low levels in the assay. NanoBRET assays involve real-time monitoring of interactions within the cell and are able to assess binding kinetics.
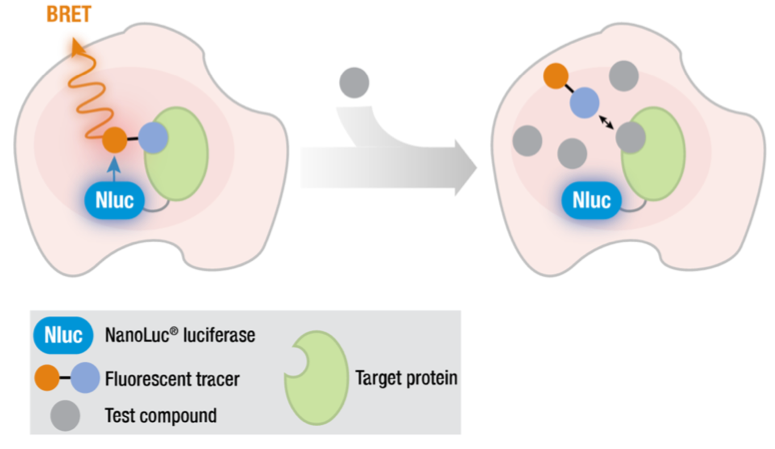
Figure. A NanoBRET target engagement assay
The key principle behind the NanoBRET assay is bioluminescence resonance energy transfer (BRET). This energy transfer occurs when a bioluminescent energy donor (e.g., the light-producing Nluc) is in close proximity to an energy acceptor (see Figure above). The acceptor is commonly a BODIPY dye incorporated into a tracer molecule that is derived from a promiscuous kinase inhibitor. When the BODIPY dye and the Nluc tagged kinase are in close proximity to each other a BRET signal is generated: energy is transferred between the donor and acceptor and, after thermal relaxation, a longer wavelength light is emitted as the observable BRET signal. As small molecule ATP-competitive inhibitors are titrated into the system, the tracer undergoes competitive binding displacement with a concomitant loss of BRET signal.
Working closely with Matt Robers and his team at Promega, we have configured NanoBRET assays for the majority of the human kinases. These assays are being used routinely at the SGC-UNC to support the characterization of cell active inhibitors for the dark kinases and for the optimization of kinase chemical probes.
SGC-UNC scientists are posting regular updates on their research on the Open Lab Notebook site: https://openlabnotebooks.org

