Resources

Read more in our perspective in SLAS Discovery: A Perspective on Extreme Open Science: Companies Sharing Compounds without Restriction. David H. Drewry, Carrow I. Wells, William J. Zuercher, and Timothy M. Willson.
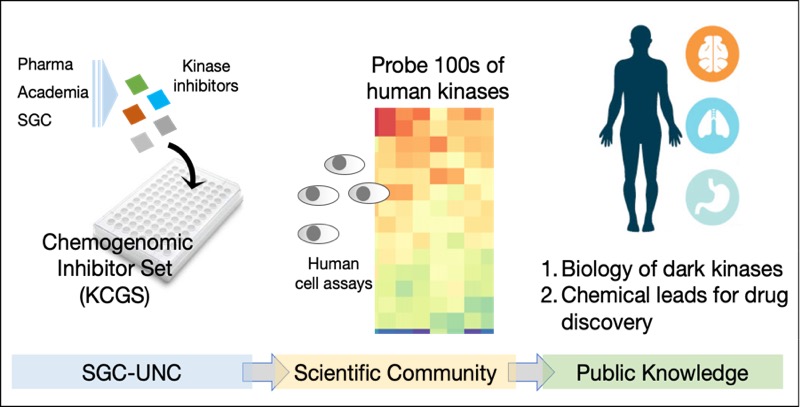
The human genome contains hundreds of dark kinases that are poorly characterized and for which little is known about their role in human disease. SGC-UNC scientists have championed a method of studying the biology of dark kinases, called chemogenomics. The approach utilizes ATP-competitive kinase inhibitors that show cross-activity on multiple kinases. The most useful ‘narrow-spectrum’ inhibitors have activity on 2 to 10 kinases. SGC-UNC scientists have assembled a large matrix of these narrow-spectrum kinase inhibitors into a chemogenomic set to probe the cell biology of the dark kinases.
Our first chemogenomic set contained 367 small molecule kinase inhibitors that were previously published by chemists at GSK. The set was named the Published Kinase Inhibitor Set (PKIS). It was carefully selected to maximize chemical and biological diversity of the inhibitors. Broad profiling at NanoSyn and the SGC Oxford showed that PKIS had activity across over 150 human kinases. PKIS was made available to the scientific community as a resource to study kinase biology and to uncover potential new targets for drug discovery. To date over 50 peer-reviewed papers have been published that report on the use of PKIS in the biomedical research.
Building on this success, a second chemogenomics set of kinase inhibitors from GSK, Takeda, and Pfizer was assembled as PKIS2. This set contained 645 inhibitors and included many additional chemotypes that were not represented in the original set. PKIS2 was profiled using the DiscoverX KINOMEscan affinity capture technology and shown to index 250 human kinases. PKIS2 was also made widely available to the scientific community. Feedback from scientists who received PKIS or PKIS2 was collected in a survey that the showed the value of these chemogenomic sets as tools to uncover new kinase biology.
Detailed analysis of the activity profiles of PKIS and PKIS2 has allowed the design of a new Kinase Chemogenomic Set (KCGS) with improved coverage of the human kinome. Importantly, KCGS contains only potent narrow spectrum kinase inhibitors thereby facilitating the annotation of phenotypic data and hypothesis generation. KCGS is now available from the SGC.
The Kinase Chemogenomic Set (KCGS) is a collection of narrow spectrum small molecule kinase inhibitors that has been assembled by the SGC-UNC to study the biology of dark kinases. KCGS is the most diverse and highly annotated publicly-available collection of kinase inhibitors. Version 1.0 of the set contains 188 kinase inhibitors sourced from eight pharma companies and academic laboratories with potent activity on 211 human kinases. Each inhibitor has been cross-screened across hundreds of kinases and only those meeting strict selectivity criteria are included in the set.
Kinase Coverage
The list of kinases covered by the inhibitors in KCGS v1.0 and their distribution across the human kinome:
AAK1 ABL1 ACVR1 ACVR1B ACVRL1 AKT1 AKT2 ALK ATM ATR AURKA AURKB AURKC BLK BMP2K BMPR1A BMPR1B BMPR2 BMX BRAF BRSK1 BRSK2 BTK BUB1 CAMKK1 CAMKK2 CASK CDK1 CDK2 CDK3 CDK4 CDK7 CDK8 CDK9 CDK12 CDK13 CDK19 CDKL2 CDKL5 CHEK1 CHEK2 CIT CLK1 CLK2 CLK3 CLK4 CSF1R CSNK1A1 CSNK1D CSNK1E DAPK3 DCLK3 DDR1 DYRK1A DYRK1B DYRK2 EGFR EIF2AK3 EPHA2 EPHA6 EPHB4 EPHB6 ERBB2 ERBB3 ERBB4 ERN1 FER FES FGFR1 FLT1 FLT3 FLT4 FRK GAK GSG2 GSK3A GSK3B HCK HIPK1 HIPK2 HIPK3 HIPK4 ICK IGF1R IKBKB IKBKE INSR INSRR IRAK1 JAK1 JAK2 JAK3 KDR KIT LATS1 LCK LIMK2 LRRK2 LTK LYN MAP2K1 MAP2K3 MAP2K4 MAP2K5 MAP3K2 MAP3K5 MAP3K7 MAP3K11 MAP3K19 MAP3K20 MAP4K2 MAP4K4 MAPK1 MAPK3 MAPK6 MAPK7 MAPK8 MAPK9 MAPK10 MAPK11 MAPK14 MAPK15 MAPKAPK2 MAPKAPK5 MARK3 MELK MET MINK1 MKNK1 MKNK2 MTOR MUSK MYLK MYLK2 NEK2 NEK3 NEK5 NEK6 NEK9 NLK NTRK1 NTRK2 NTRK3 NUAK1 NUAK2 PAK1 PDGFRA PDGFRB PDPK1 PHKG1 PI4KB PIK3C2B PIK3C2G PIK3C3 PIK3CA PIK3CB PIK3CD PIK3CG PIM1 PIM2 PIM3 PIP4K2C PIP5K1C PKN2 PLK1 PLK2 PLK3 PRKAA2 PRKCD PRKCE PRKCH PRKCQ PRKD1 PRKD2 PRKD3 PRKX PTK2 PTK2B PTK6 RAF1 RET RIOK1 RIOK2 RIPK1 RIPK2 RIPK3 ROCK1 ROCK2 RPS6KA1 RPS6KA2 RPS6KA3 RPS6KA4 RPS6KA5 RPS6KA6 RPS6KB1 SIK1 SRC SRMS STK10 STK16 STK17A STK17B STK36 SYK TAOK3 TBK1 TEK TGFBR1 TNK1 TNK2 TNNI3K TTK TYK2 YES1 ZAP70

For additional information see the KCGS FAQ
KCGS Availability
The current set is available to any investigator at a non-for-profit organization for an access fee of $3100 that off-sets the resynthesis cost of the small molecule inhibitors. Requestors receive:
- A copy of KCGS in 384-well plates as 1 µL of 10 mM DMSO solution
- A spreadsheet that lists chemical structure, target kinase, and literature references for each inhibitor
- 5 x 1 µL of 10 mM DMSO cherry picks upon request
- Access to the full kinase selectivity data for each inhibitor
- KCGS v1.0 can be requested through the following link.
A list of investigators that have received a copy of KCGS can be found here.
KCGS Resynthesis
To ensure the continued availability of KCGS, we have embarked on a project to resupply all of the inhibitors in the set. Current status of the resynthesis project is shown below:
Last Updated 4-26-19
Acknowledgment
Funding to support the assembly and annotation of KCGS was provided by the UNC Eshelman Institute for Innovation, the NIH Illuminating the Druggable Genome (1U24DK116204-01), and the SGC. Kinase inhibitors were made available by AbbVie, Bayer, Boehringer Ingelheim, GlaxoSmithKline, Merck Sharp & Dohme, Pfizer, Takeda, AstraZeneca, and Vertex. Additional kinase inhibitors were provided by Nathanael Gray (Dana Farber Cancer Institute) and Julian Blagg (Cancer Research UK). Screening assays were supported by Promega and Eurofins-DiscoverX. Funding to support resynthesis of KCGS has been provided by the University Cancer Research Fund, UNC Eshelman Institute for Innovation, the Office of the UNC Vice Chancellor for Research, the UNC School of Medicine, and the SGC.
A scientific manuscript and supporting information describing KCGS with details of the kinases covered and its use in cell-based assays is available for download from bioRxiv.
Kinase Inhibition Data
A simple Excel spreadsheet listing the kinases covered by each inhibitor, its physical properties, and chemotype is available here. This spreadsheet can be used to rapidly analyze screening data to identify candidate kinases for follow up experiments.
A single downloadable file of with the full inhibition profiles is not currently available, since the data are not in a uniform format. Most of the compounds have been profiled across 401 human kinases using the DiscoverX KinomeSCAN technology, with KD’s generated for kinases that were inhibited by 90% at 1.0 µM. However, 42 compounds were selected using data available from the 230 kinase NanoSYN panel collected at 0.1 and 1.0 µM. An additional 21 compounds utilized published data from a range of large kinase panels that was collected in multiple assay formats and at different concentrations. We are in the process of collecting DiscoverX KinomeSCAN data for these 63 compounds and will make a uniform KCGS dataset available for download as soon as it is available. In the interim, the kinase inhibition profile of each individual compound using the mixed assay formats can be accessed at the on-line database Random Acts of Kinase.
Chemical Structures
Chemical structures are provided in the Excel spreadsheet in SMILES notation. The chemical structures are also available in a fully structure searchable format as a Datawarrior file.
Information on how to download the free cheminformatics program Datawarrior can be found here.
KCGS Plate Map
KCGS is shipped in a 384-well microtiter plate. A physical copy of the plate map with compound identifiers is available here. For recommended dilution protocols see the FAQ.
The first release of KCGS is available for distribution. The fee for KCGS v1.0 is $3,100, which helps to defray resynthesis costs to make KCGS a sustainable resource for all research labs.
KCGS is supplied in 1µL aliquots of a 10 mM stock solution in DMSO of 187 compounds. You will have access to the chemical structure and to kinome-wide selectivity data for each compound, and you can receive 5µL of up to 5 compounds for follow-up screening. For additional cherry picks, cost is determined by quantity requested.
For UNC Researchers:
Requests for KCGS can be submitted through Infoporte CORES. You will be required to provide a chartfield string for the cross charge.
For non-UNC Researchers:
Requests for KCGS can be submitted through Ximbio. You will be required to complete an institutional material transfer agreement.
All Recipients of KCGS must abide by the material trust agreement below:
The Structural Genomics Consortium (SGC) accelerates research in new areas of human biology and drug discovery (1) by ensuring that its Materials and research outputs derived from them are available in the public domain to the scientific community on a pre-competitive basis, without restrictions based on patents or other forms of intellectual property, and (2) by creating an open collaborative network of scientists around the world who are committed to this principle.
The Material is therefore entrusted to you for the benefit of:
(1) the SGC and its members (including your Establishment where applicable),
(2) individual members of the scientific research community whose work may be advanced by open access to the Material and to research results and data outputs arising from your use of the Material, and
(3) individual members of the general public whose health may be ameliorated or improved as a result of your work with the Material, and the work of other members of the scientific community who enjoy free and open access to the Material (together, the “Beneficiaries”).
Your obligations as trustee as described to the right do not extend to new compounds that may be generated or synthesized in connection with your use of the Material (the ‘Modifications’). Modifications mean changes to the chemical structure of the Material, including rearrangements of covalent bonds or additions or removals of atoms, residues, or moieties connected by covalent bonds, except ester forms. Modifications do not include resolved stereoisomers, salt forms, hydrates, solvates, polymorphs, complexes, or other non- covalent derivatives of the Material, nor do they include methods of synthesis, uses, formulations, or dosages of the Material.
BY ACCEPTING THE MATERIAL, YOU AGREE TO HOLD THE MATERIAL IN TRUST FOR THE BENEFIT OF THE BENEFICIARIES, AND, IN YOUR CAPACITY AS A TRUSTEE, YOU AGREE TO THE FOLLOWING OBLIGATIONS AND RESPONSIBILITIES WITH RESPECT TO THE MATERIAL: *
1. YOU WILL NOT FILE AN APPLICATION FOR PATENT, NOR ACTIVELY SEEK ANY OTHER FORM OF INTELLECTUAL PROPERTY PROTECTION, CLAIMING OR COVERING THE MATERIAL OR ITS PHYSICAL FORMS, METHODS OF SYNTHESIS, USES, FORMULATIONS, OR DOSAGES, NOR WILL YOU PERMIT OTHERS UNDER YOUR DIRECT SUPERVISION TO SEEK, NOR WILL YOU ASSIST OTHERS IN SEEKING, ANY SUCH PATENTS OR OTHER INTELLECTUAL PROPERTY PROTECTIONS. IF ANY INVENTION OR DISCOVERY PERTAINING TO THE MATERIAL, WHETHER PATENTABLE OR NOT, IS FIRST CONCEIVED OR REDUCED TO PRACTICE BY YOU, YOUR ESTABLISHMENT, OR THOSE UNDER YOUR DIRECT SUPERVISION, DURING RESEARCH USING THE MATERIAL, YOU AGREE NOT TO ENFORCE ANY RIGHTS THAT YOU MAY HAVE IN THE INVENTION OR DISCOVERY AGAINST THE SGC OR ITS MEMBERS, AND YOU HEREBY GRANT TO THE SGC AND ITS MEMBERS A NON-EXCLUSIVE, IRREVOCABLE, PERPETUAL, WORLD-WIDE, SUB-LICENSABLE, AND ROYALTY-FREE RESEARCH USE LICENSE UNDER ANY SUCH RIGHTS.
2. YOU AGREE TO PLACE THE RESULTS OF THE RESEARCH PERFORMED WITH THE MATERIAL BY YOU, YOUR ESTABLISHMENT, AND THOSE UNDER YOUR DIRECT SUPERVISION, IN THE PUBLIC DOMAIN, BY PUBLISHING RESULTS IN ONE OR MORE OPEN ACCESS JOURNALS AND BY DEPOSITING THE UNDERLYING DATA IN A PUBLIC ACCESS REPOSITORY OR, IF NOT FEASIBLE, IN AN INSTITUTIONAL REPOSITORY ACCESSIBLE TO ACADEMIC RESEARCHERS, WITHIN ONE YEAR OF PUBLICATION.
3. YOU AGREE THAT YOU, YOUR ESTABLISHMENT, AND THOSE UNDER YOUR DIRECT SUPERVISION WILL ACKNOWLEDGE THE PROVIDERS LISTED IN APPENDIX A AS THE SOURCE OF THE MATERIAL IN ALL PUBLICATIONS RESULTING FROM, AND IN ALL PRESENTATIONS IN SCIENTIFIC FORA REGARDING, USE OF THE MATERIALS.
4. YOU AGREE TO PROVIDE THE SGC WITH A BRIEF UPDATE ON YOUR PROGRESS WITH THE MATERIALS AND A REPORT BRIEFLY DESCRIBING THE RESULTS OF THE RESEARCH UNDERTAKEN WITH THE MATERIAL, UPON CONCLUSION OF SUCH RESEARCH. YOU AGREE TO PROVIDE A COPY OF ALL PROPOSED PUBLICATIONS TO THE SGC PRIOR TO SUBMISSION.
5. YOU AGREE TO HAVE YOUR NAME AND INSTITUTION LISTED AS A RECIPIENT OF KCGS ON THE SGC WEBSITE AND SGC PUBLICATIONS.
6. YOU AGREE THAT THE MATERIAL IS FOR IN VITRO LABORATORY USE ONLY AND WILL NOT BE USED IN HUMANS OR ANIMALS, WITHOUT FIRST OBTAINING WRITTEN CONSENT FROM SGC.
7. YOU AGREE TO USE THE MATERIAL IN COMPLIANCE WITH ALL APPLICABLE FEDERAL, PROVINCIAL, STATE, COUNTRY, AND LOCAL LAWS, REGULATIONS, AND ORDINANCES.
8. YOU AGREE THAT THE MATERIAL WILL BE USED FOR TEACHING OR NOT-FOR-PROFIT RESEARCH PURPOSES ONLY.
9. YOU AGREE NOT TO DISTRIBUTE THE MATERIAL TO, OR ALLOW ACCESS TO THE MATERIAL BY, ANY THIRD PARTIES WITHOUT THE WRITTEN CONSENT OF SGC.
10. IN SITUATIONS WHERE THE EU GDPR APPLIES, I UNDERSTAND AND AGREE THAT BY CHECKING THE BOX AND PRESSING THE SUBMIT BUTTON THAT I AM AUTHORIZING THIS SITE AND UNC CHAPEL HILL TO COLLECT AND PROCESS PERSONAL INFORMATION ABOUT ME FOR THE PURPOSE OF INTERACTING WITH THIS SITE AND FOR THE APPLICABLE PURPOSES LISTED BELOW: DISPLAYING A LIST OF THE RECIPIENTS OF KCGS ON THE SGC WEBSITE; RECEIVING PERIODIC UPDATES FROM THE SGC ON THE EXPANSION OR CHARACTERIZATION OF KCGS; RECEIVING REQUESTS FROM THE SGC FOR INPUT ON HOW TO IMPROVE KCGS. IN THE CASE WHERE THE EU GDPR APPLIES AND I WISH TO WITHDRAW MY CONSENT FOR MY PERSONAL INFORMATION TO BE RETAINED BY THIS SITE OR UNC-CHAPEL HILL, I UNDERSTAND THAT I WILL NEED TO CONTACT THE SITE ADMINISTRATOR AND UNC-CHAPEL HILL. MY CONSENT TO THE ABOVE PROCESSING IS INFORMED, FREELY GIVEN, FOR THE SPECIFIC PURPOSES LISTED HEREIN, AND REPRESENTS AN UNAMBIGUOUS INDICATION OF MY WISHES TO AGREE TO THE PROCESSING OF PERSONAL DATA RELATED TO ME.
What is KCGS?
The Kinase Chemogenomic Set (KCGS) v1.0 is a collection of 187 potent and selective small molecule ATP-competitive inhibitors that were produced in drug discovery programs targeting protein kinases. Each inhibitor has been profiled for binding against >200 human kinases at DiscoverX. Each inhibitor has a Kd < 100 nM for its target kinase with a selectivity index S10 < 0.04 at 1 µM. In total, KCGS indexes 215 human kinases including many poorly studied (dark) kinases. KCGS is available from the SGC in aliquots of 1 µL of a 10 mM solution in DMSO.
How do I find more information about KCGS?
An open access scientific paper and supporting information describing KCGS with details of the kinases covered and its use in cell-based assays has been published in the International Journal of Molecular Sciences https://doi.org/10.3390/ijms22020566
Where can I get the chemical structures and kinase inhibition data?
Details on how to access the data are provided here KCGS data page. The chemical structures of the inhibitors and the kinase activity data are available on our website Random Acts of Kinase.
How do I obtain KCGS?
Detailed instructions for requesting the set are posted on our web page. UNC Researchers have access to KCGS through Core Serices. All other researchers have acces to KCGS through Ximbio.
Why are you charging for KCGS?
KCGS is currently a finite resource that is available in limited supply through donation of kinase inhibitors from pharma companies and academic labs. We will use all funds raised through distribution of KCGS to finance resynthesis of the kinase inhibitors so we can continue to make it available as a public resource.
How did you decide the $3100 processing and distribution fee?
A survey of recipients of PKIS and PKIS2 indicated that the majority would have paid between $1000-$5000 for access to the sets. All funds raised through distribution of KCGS will be used to support resynthesis of the kinase inhibitors so we can provide the set for years to come.
What if I cannot afford the processing and distribution fee?
We will gladly work with you to submit grant applications to raise the funds. It is likely that the KCGS processing and distribution fee will be only a fraction of the operational cost of your proposed experiment.
How should we store KCGS upon receipt?
We recommend storing the plate between -20 and -80°C until use. Researchers are advised to avoid multiple freeze-thaw cycles with the KCGS plate to ensure best results.
What concentration range should be used when running a cellular screen using the KCGS library?
KCGS contains potent kinase inhibitors that have been profiled for selectivity at 1 μM. We recommend cellular screens be conducted no higher than 1 μM. If assay throughput can accommodate it, another option is to conduct the initial screening at 2-3 different concentrations; for example 10 nM, 100 nM, and 1 μM.
How are KCGS compounds solubilized?
All KCGS compounds are dissolved in DMSO and dispensed as 1 μL of a 10 mM stock solution.
What kind of plate is KCGS dispensed into?
KCGS is dispensed in a Greiner #781280 V-bottom polypropylene 384-well plate as 1 μL of a 10 mM stock solution.
What is the total capacity of the wells in the KCGS plate?
The capacity of the KCGS wells is 130 μL. It is recommended to fill up to 100 μL if automation is involved.
Is there a recommended dilution protocol for the KCGS plate?
When ready to dilute the plates, we recommend bringing the KCGS plate to room temperature, spinning down all liquid, diluting and agitating. We recommend diluting with DMSO. For example, adding 9 μL of DMSO to the 1 μL of 10 mM stock will yield 10 μL of 1 mM stock in your KCGS plate. A 1000-fold dilution of the stock plate in assay media will yield 10 mL of a 1 μM solution containing 0.1% DMSO.
How much can I request?
We dispense KCGS as 1 µL of a 10 mM solution in DMSO. As KCGS is currently a limited resource and you require more than the standard dispensement we will work with you to define the minimal volume necessary to enable the proposed experiments and decide if we can meet that need.
Is there space for controls on the KCGS plate?
The first two and last two columns of every plate are left intentionally blank for control wells when plating KCGS.
Which compounds are contained in my plate?
Physical and electronic plate maps are included with every shipment. We will be providing plate maps on our website that people can access using the plate number they receive as well.
What is known about the stability of compounds in KCGS?
The compounds in KCGS are not known to have any light or air sensitivities. A QC plate was analyzed prior to dispensing the KCGS plates. However, researchers are advised to avoid multiple freeze-thaw cycles with the KCGS plate to ensure best results.
Does SGC-UNC have general toxicity data for the KCGS library?
Details of the general toxicity at 10 µM inhibitor concentration in HeLa cells can be found in the open access paper https://doi.org/10.3390/ijms22020566 The effect of the inhibitors at 1.0 µM concentration on growth rate across 17 cell lines is also detailed in the manuscript.
What is the kinome coverage of KCGS?
Using data collected across 401 human kinases, KCGS covers 54% of the screenable kinome. Each inhibitor has a Kd <100 data-preserve-html-node=”true” nM for its target kinase with a selectivity index S10 <0.04 data-preserve-html-node=”true” at 1 µM. There are no highly promiscuous kinase inhibitor in KCGS.
How do I share data generated with KCGS?
The data can be published in scientific literature, shared by emailing it to us, or by connecting with a data repository such as ChEMBL.
Do I need to include SGC-UNC scientists on any publications that arise from use of the set?
We do not feel that simple supply of compounds justifies inclusion as coauthors. If we add something more to a collaborative effort, please use normal academic standards for coauthorship. However, all publications should identify the source of KCGS as the SGC-UNC and you should send a copy of the citation to us upon publication. We will post a list of all publications arising from the use of KCGS on our website.
What do I need to put in the Acknowledgments section of a publication?
We recommend using the following text “KGCS was supplied by the SGC-UNC and contains kinase inhibitors that were made available by GlaxoSmithKline, Takeda, Pfizer, Boehringer Ingelheim, Vertex, Harvard University, Cancer Research UK, and the SGC.”
What are the requirements for using the set?
Recipients of KCGS must use the set within 90 days. Recipients of KCGS will be identified on our website. You must communicate results to us and make all data derived from use of the sets publically available within a reasonable amount of time.
I have some interesting results. Can I get additional quantities of compounds of interest?
We try to supply additional quantities compounds when possible from our internal supplies, but these are often limited. However, we will always provide options for obtaining additional quantities of the compounds through resynthesis.
I have some interesting results. Can I get analogs of compounds of interest?
We have analogs of many of the kinase inhibitors in KCGS that may be useful for establishing structure-activity relationships around a kinase of interest.
Where can I get the chemical structures and kinase inhibition data?
Details on how to access the data are provided here
I want to pursue an in vivo experiment with a molecule from KCGS. Is this possible?
KCGS is formatted for screening in cells and little is known about the pharmacokinetic properties of the individual inhibitors. If you plan to use a KCGS compound in animals, evidence of an appropriate institutional animal care and use committee (IACUC or equivalent) protocol approval is required before we can consider a request of this nature. Quantities of individual compounds maybe severely limited and ultimately preclude supply for an in vivo study.

SGC-UNC is contributing chemical tools and NanoBRET assays to the IDG Drug and Resource Generation Center for Kinases led by Gary Johnson.
IDG Chemical Tools are cell active inhibitors for dark kinases on the IDG target list. Each inhibitor shows IC50 < 1 µM in a NanoBRET assay for its target dark kinase and also demonstrates a narrow spectrum of activity across a large panel of kinases (S10 < 0.05 at 1 µM).
The current list of IDG chemical tools with links to their supporting data is available on the Dark Kinase Knowledgebase.
Chemical tools that are not commercially available can be requested from the SGC-UNC.
IDG tool molecules are compounds that we believe are important for investigating the function of understudied kinases on the Illuminating the Druggable Genome list. Compounds synthesized or provided by the SGC-UNC can be shipped to researchers upon completion of the click-through MTA (below) and payment through our infoporte site.
The Structural Genomics Consortium (SGC) accelerates research in new areas of human biology and drug discovery
(1) by ensuring that its Materials and research outputs derived from them are available in the public domain to the scientific community on a pre-competitive basis, without restrictions based on patents or other forms of intellectual property, and (2) by creating an open collaborative network of scientists around the world who are committed to this principle.
The Material is therefore entrusted to you for the benefit of
(1) the SGC and its members (including your Establishment where applicable),
(2) individual members of the scientific research community whose work may be advanced by open access to the Material and to research results and data outputs arising from your use of the Material, and
(3) individual members of the general public whose health may be ameliorated or improved as a result of your work with the Material, and the work of other members of the scientific community who enjoy free and open access to the Material (together, the “Beneficiaries”).
Your obligations as trustee as described to the right do not extend to new compounds that may be generated or synthesized in connection with your use of the Material (the ‘Modifications’). Modifications mean changes to the chemical structure of the Material, including rearrangements of covalent bonds or additions or removals of atoms, residues, or moieties connected by covalent bonds, except ester forms. Modifications do not include resolved stereoisomers, salt forms, hydrates, solvates, polymorphs, complexes, or other non-covalent derivatives of the Material, nor do they include methods of synthesis, uses, formulations, or dosages of the Material.
Request IDG Tools
Submit a Request for KCGS

